
Third, a “majority-voting” method determines the level Of individual experiments and the correlation between experiments. (TW) method that computes a weighted sum of counts for a given region, where the weights account for both the TSS enrichments First, a “naïve Bayes” method computes a score that combines the ChIP signals in different experiments, whichĪre thresholded and weighted according to how well they perform on a set of known promoters. To this end, we have implemented four complementary methods to integrate the compendium of ENCODE transcriptional regulatoryĮlement data. Together, as a modest signal that is reproduced across a number of experiments can become much more significant than it would These shortcomings can be overcome when a number of experiments are analyzed Set is often arbitrary, and individual data points near the threshold can be easily misclassified depending on whether theĮmphasis is placed on specificity or sensitivity. Therefore, invoking a threshold for calling a site bound or unbound by a transcription factor in an individual data In the case of ChIP-chip, a discreet biochemical event (e.g., histone modification) is usually not reflected as a binary experimental Significant analytical challenges with microarray-based functional genomics is the continuous nature of the data. That evaluated the data as a whole in a quantitative manner rather than studying each data set individually. Throughout the ENCODE-defined regions independent of mRNA to genomic DNA sequence alignments. With this set of transcriptional regulatory element data, we aimed to map transcriptional promoters and regulatory regions This compendium of data thus providesĪn unprecedented collection of experimental observations characterizing transcription start sites (TSSs) and their associated Initiation, such as TAF1, methylation of lysine 4 on histone H3, and RNA polymerase II. Many of these experiments were conducted on factors known by previous studies to mark sites of transcription Immunoprecipitation combined with genomic microarrays (ChIP-chip) or tag sequencing. The majority of these data sets measure transcription-factor binding and histone modifications using the technique of chromatin Over 150 independent experiments were conducted to characterize transcriptional regulatory elements in human cell lines. The pilot phase of The ENCODE Project Consortium has generated a large volume and variety of functional genomics data ( The ENCODE Project Consortium 2004, 2007). Results suggest that there are at least 35% more functional promoters in the human genome than currently annotated. We also performed 5′-RACEĮxperiments on 62 novel regions, and 76% of the regions were associated with the 5′-ends of at least two RACE products. We tested 163 such regions for promoter activity in four cell lines using transient transfectionĪssays, and 25% of them showed transcriptional activity above background in at least one cell line. These novel regions may regulate unannotated transcripts or may represent new alternative transcription The majority of the predicted regions are >2 kb away from the 5′-ends of previously annotated humanĬDNAs and hence are novel. Interestingly, a significant portion of the regions overlaps with annotated 3′-UTRs, suggesting that some of them might regulateĪnti-sense transcription.
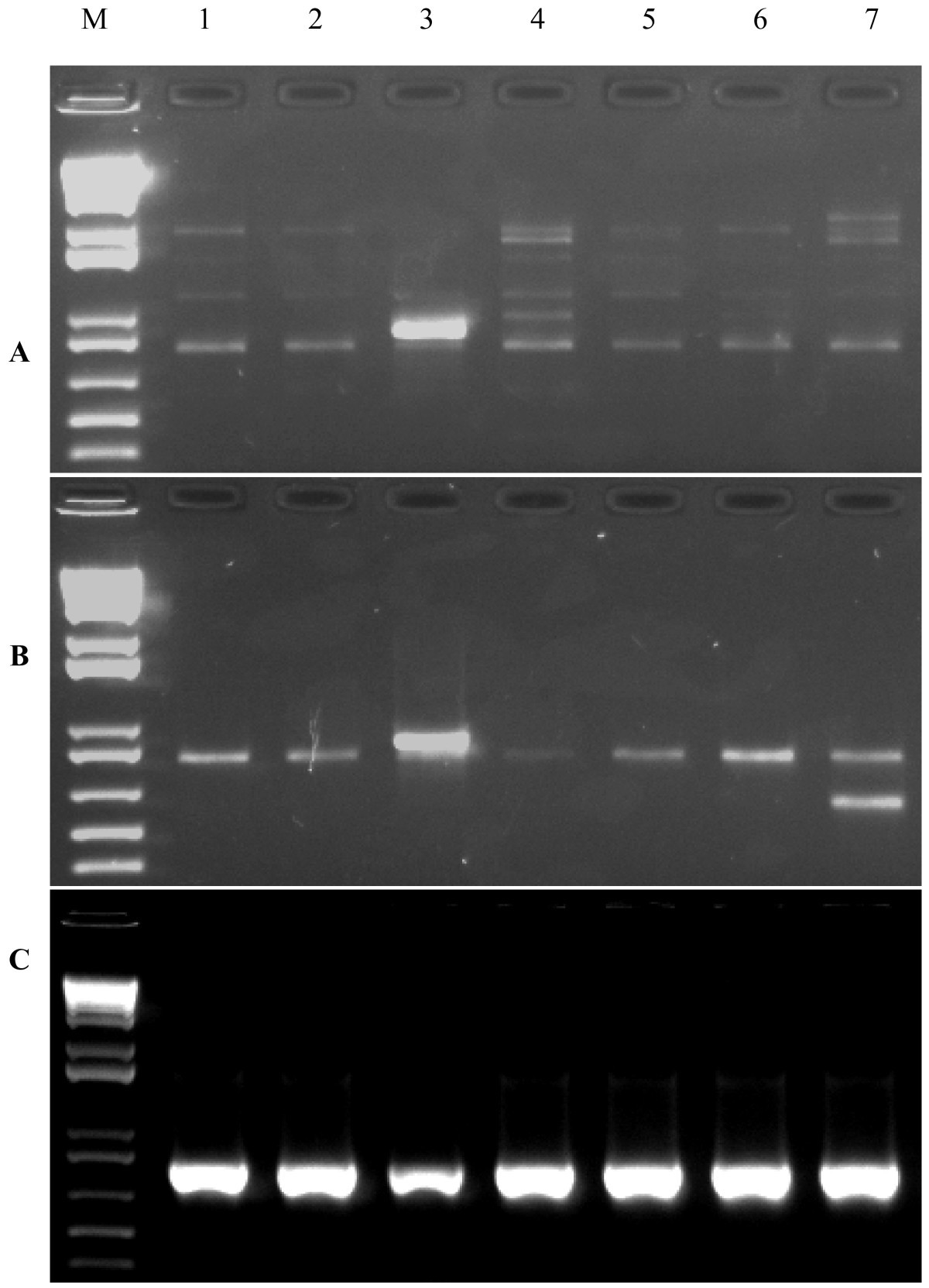
Predicted regions tend to localize to highly conserved, DNase I hypersensitive, and actively transcribed regions in the genome. Roughly 47% of the regions were unique to one method, as each method makes different assumptions about the data. They collectively predicted 1393 regions. Methods to integrate 129 sets of ENCODE-wide chromatin immunoprecipitation data. In order to identify regulatory regions, we developed four computational

The regulation of transcriptional initiation in the human genome is a critical component of global gene regulation, but aĬomplete catalog of human promoters currently does not exist.


 0 kommentar(er)
0 kommentar(er)
